Imagine walking into a crowded comic‑con where everyone is in costume, talking at once, and you’re supposed to find your friend dressed as one very specific superhero. That chaos? That’s your crude protein mixture. In swoops affinity chromatography, the over‑enthusiastic super‑fan with a VIP list and a velvet rope, grabbing only the molecules with the right “name tag” and letting everyone else wander off. Suddenly, the room is quieter, but there are still sidekicks, bodyguards, and a few random photobombers hanging around your hero. That’s when size exclusion chromatography steps in as the sensible event organizer, sorting everyone by size and gently escorting them to the right exit. Together, these two methods turn biochemical bedlam into an orderly, almost polite separation.
What each method actually does
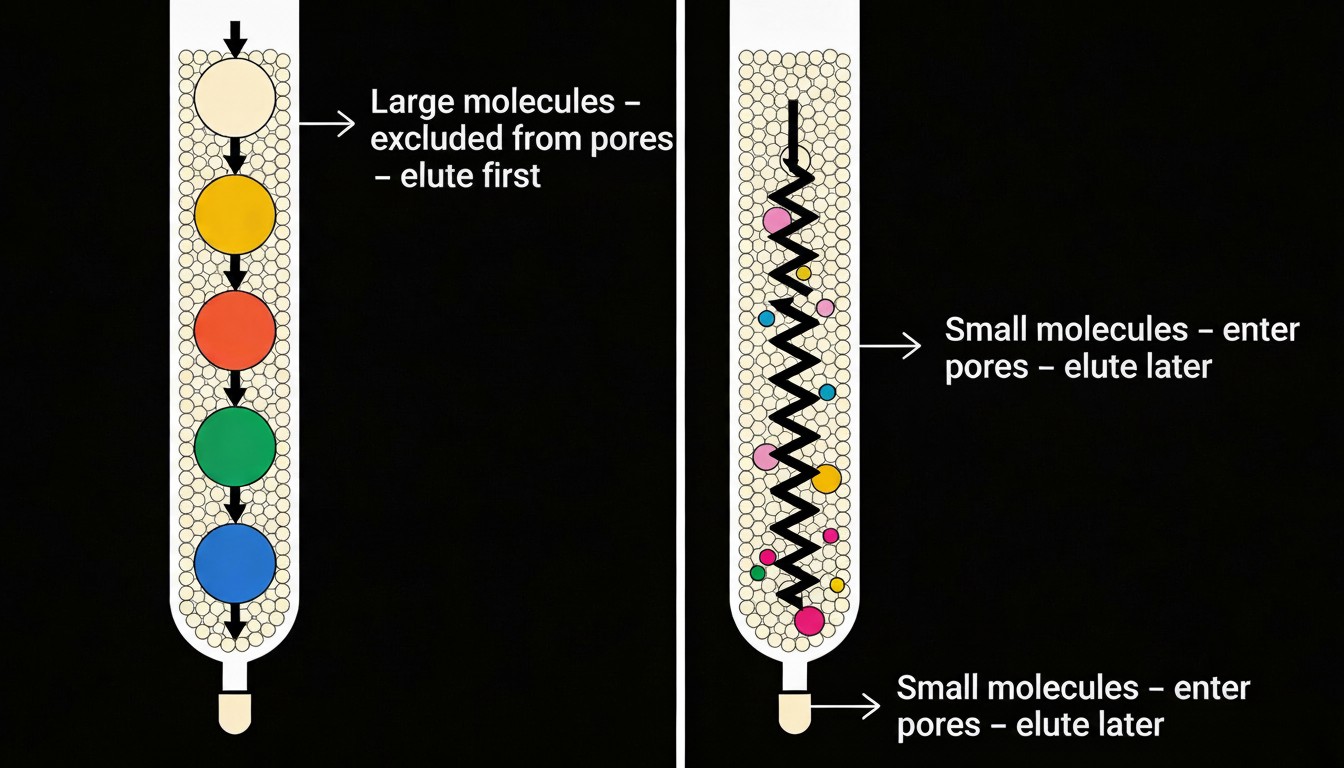
Size exclusion chromatography (SEC, also called gel filtration) separates molecules based primarily on hydrodynamic size as they pass through a column packed with porous beads. Large molecules cannot enter the pores and elute first, while smaller molecules penetrate the pores, take a longer path, and elute later, all without strong binding to the stationary phase
This illustration roughly shows what happens during size exclusion chromatography. The stable phase is tuned to hold a specific sized molecule, and everything else passes through. Affinity chromatography, in contrast, relies on specific and reversible interactions between a target molecule and an immobilized ligand such as an antibody, metal chelate, lectin, or biotin‑binding protein. The target binds to the ligand while most other components wash straight through, and is then eluted by changing conditions like pH, ionic strength, or adding a competing ligand
How they are similar (and different)
Both techniques use columns packed with beads and a flowing mobile phase, and both are workhorses for protein purification in research and bioprocessing. Yet SEC is largely “non‑interactive” and gentle, preserving native structure, while affinity chromatography is deliberately interactive and often achieves orders‑of‑magnitude purification in a single step.
In practice, SEC is excellent for polishing: removing aggregates, exchanging buffers, and fractionating by size; affinity is superb for capturing a specific species from a complex mixture, especially when a well‑behaved ligand–target pair is available. Together, they create a workflow where affinity does the heavy lifting and SEC cleans up the fine details such as monomer versus aggregate or different complex stoichiometries
Where combining them really shines
A classic use case is monoclonal antibody work: Protein A affinity chromatography captures IgG from cell culture, and SEC then resolves monomeric antibody from dimers, higher‑order aggregates, and fragments. Similar combined strategies are used for extra. Then hand your newly purified friend off to SEC for finishing school: match the column’s fractionation range to the molecular weight you care about, keep sample volume around 5–10% of total column volume, and avoid overloading to preserve resolution. Because SEC is gentle and primarily size‑based, it will let you separate monomers from aggregates or resolve complexes of different sizes without strong secondary interactions, making your final pool much more “publication‑ready.” cellular vesicles, where affinity or “bind–elute” SEC media enrich vesicles and conventional SEC further removes protein and RNA contaminants
Analytical methods push this even further: affinity‑resolved SEC coupled to mass spectrometry can probe specific interactions (for example, antigen–antibody or Fc receptor binding) while simultaneously characterizing size distribution and structural variants. In quality control settings, coupled affinity–SEC HPLC can monitor both binding behavior (via the affinity step) and aggregation state (via SEC) in the same workflow, providing a richer picture of product quality
A friendly design checklist
When building an affinity‑plus‑SEC workflow, it helps to think like an engineer with a sense of humor. First, choose an affinity ligand that truly loves your target more than everything else in the lysate—Protein A for IgG, metal chelates for His‑tags, or lectins for specific glycoforms are common options. Second, design elution conditions that release the target without wrecking it; harsh pH may be fine for a robust enzyme but disastrous for a delicate complex you plan to analyze by SEC
Here are two great industry resources to look at if you’re going to try it:
Cube Biotech resource page lays out the system beautifully, and their tables show a great way to understand what to choose at what stages.BioRad has a more exhaustive chart of matrix materials you might choose from.
Quick side‑by‑side snapshot
Feature Size exclusion chromatography Affinity chromatography
Main separation basis
Hydrodynamic size in porous beads.
Specific ligand–target binding on immobilized ligands.
Typical role
Polishing, desalting, aggregate removal.
Capture and major enrichment from complex mixtures.
Interaction strength
Minimal, largely non‑interactive and gentle.
Strong but reversible specific interactions.
Resolution vs. mixture
Good for size differences within a defined range.
Very high for molecules that bind; poor for non‑binders.
Common pairing
Often used after affinity or ion exchange as final step.
Frequently used as first step before SEC polishing.
When these two techniques are combined thoughtfully, the result is a robust, modular purification pipeline that can turn chaotic biological soup into clean, well‑behaved samples ready for biophysical characterization, omics workflows, or scaled manufacturing. For an academic lab or a small R&D shop, mastering this combo is one of the fastest ways to upgrade both data quality and day‑to‑day sanity on the bench.
{21495:PWBGF9AM};{21495:WZA8I8AK};{21495:RT4XJVNM};{21495:9J7HLWKI},{21495:UE2BXCY9},{21495:U6XAGSRZ};{21495:XG7ZVESZ};{21495:YV9LWY3C}
apa
default
asc
0
13060
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3Afalse%2C%22meta%22%3A%7B%22request_last%22%3A550%2C%22request_next%22%3A50%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%229F28B8Q3%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22anubha%22%2C%22parsedDate%22%3A%222024-11-11%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3Banubha.%20%282024%2C%20November%2011%29.%20Lab%20Work%20Made%20Easy%3A%20Choosing%20the%20Right%20Chemistry%20Glassware.%20%26lt%3Bi%26gt%3BScientific%20Lab%20Equipment%20Manufacturer%20and%20Supplier%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.shivsons.com%5C%2Flab-work-made-easy-choosing-the-right-chemistry-glassware%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.shivsons.com%5C%2Flab-work-made-easy-choosing-the-right-chemistry-glassware%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Lab%20Work%20Made%20Easy%3A%20Choosing%20the%20Right%20Chemistry%20Glassware%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22anubha%22%7D%5D%2C%22abstractNote%22%3A%22Find%20the%20right%20chemistry%20glassware%20for%20your%20lab.%20Simplify%20experiments%20and%20boost%20accuracy%20with%20essential%20tips%20for%20lab%20work%20made%20easy.%22%2C%22blogTitle%22%3A%22Scientific%20Lab%20Equipment%20Manufacturer%20and%20Supplier%22%2C%22date%22%3A%222024-11-11T04%3A55%3A33%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.shivsons.com%5C%2Flab-work-made-easy-choosing-the-right-chemistry-glassware%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-26T17%3A39%3A32Z%22%7D%7D%2C%7B%22key%22%3A%225E8EW9NC%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22The4%22%2C%22parsedDate%22%3A%222026-01-29%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BThe4.%20%282026%2C%20January%2029%29.%20%26lt%3Bi%26gt%3BA%20list%20of%20glassware%20equipment%20for%20high%20school%20chemistry%20lab.%20-%20Science%26lt%3B%5C%2Fi%26gt%3B.%20Science%20Equip.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.scienceequip.com.au%5C%2Fblogs%5C%2Fnews%5C%2Fa-list-of-glassware-equipment-for-high-school-chemistry-lab%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.scienceequip.com.au%5C%2Fblogs%5C%2Fnews%5C%2Fa-list-of-glassware-equipment-for-high-school-chemistry-lab%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22A%20list%20of%20glassware%20equipment%20for%20high%20school%20chemistry%20lab.%20-%20Science%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22The4%22%7D%5D%2C%22abstractNote%22%3A%22In%20the%20article%20we%20take%20a%20closer%20look%20at%20some%20of%20the%20most%20common%20glassware%20equipment%20utilised%20by%20high%20school%20chemistry%20lab%20today.%22%2C%22date%22%3A%222026-01-29%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.scienceequip.com.au%5C%2Fblogs%5C%2Fnews%5C%2Fa-list-of-glassware-equipment-for-high-school-chemistry-lab%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-26T17%3A39%3A24Z%22%7D%7D%2C%7B%22key%22%3A%22KSL995K3%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22says%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3Bsays%2C%20G.%20johnson.%20%28n.d.%29.%20%26lt%3Bi%26gt%3BBeakers%20vs.%20Flasks%3A%20The%20Pros%20and%20Cons%20of%20Frequently%20Used%20Lab%20Glassware%26lt%3B%5C%2Fi%26gt%3B.%20Lab%20Pro%20Inc.%20Retrieved%20February%2026%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Flabproinc.com%5C%2Fblogs%5C%2Flaboratory-equipment%5C%2Fbeakers-vs-flasks-the-pros-and-cons-of-frequently-used-lab-glassware%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Flabproinc.com%5C%2Fblogs%5C%2Flaboratory-equipment%5C%2Fbeakers-vs-flasks-the-pros-and-cons-of-frequently-used-lab-glassware%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Beakers%20vs.%20Flasks%3A%20The%20Pros%20and%20Cons%20of%20Frequently%20Used%20Lab%20Glassware%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gary%20johnson%22%2C%22lastName%22%3A%22says%22%7D%5D%2C%22abstractNote%22%3A%22Find%20out%20%5Cu201cwhat%20is%20a%20beaker%20and%20a%20flask%5Cu201d%2C%20%5Cu201cwhat%20are%20they%20used%20for%20in%20labs%5Cu201d.%20Compare%20beakers%20and%20flasks%20to%20choose%20the%20right%20one%20for%20precision%20and%20consistency.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Flabproinc.com%5C%2Fblogs%5C%2Flaboratory-equipment%5C%2Fbeakers-vs-flasks-the-pros-and-cons-of-frequently-used-lab-glassware%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-26T17%3A16%3A01Z%22%7D%7D%2C%7B%22key%22%3A%22FHRVBIGC%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BUnderstanding%20Laboratory%20Glassware%20Classifications%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20Globe%20Scientific.%20Retrieved%20February%2026%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fglobescientific.com%5C%2Fblogs%5C%2Fblog%5C%2Funderstanding-laboratory-glassware-classifications%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fglobescientific.com%5C%2Fblogs%5C%2Fblog%5C%2Funderstanding-laboratory-glassware-classifications%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Understanding%20Laboratory%20Glassware%20Classifications%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22As%20a%20busy%20lab%20professional%2C%20you%20need%20to%20have%20confidence%20that%20the%20glassware%20in%20your%20lab%20is%20reliable%20and%20of%20high%20quality.%20You%20don%5Cu2019t%20have%20time%20to%20worry%20about%20how%20your%20glassware%20will%20perform%20in%20measurement%20settings%5Cu2014you%20need%20the%20peace%20of%20mind%20that%20it%20can%20compete%20with%20the%20best%20products%20on%20the%20market.%20Luckily%2C%20product%20manufac%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fglobescientific.com%5C%2Fblogs%5C%2Fblog%5C%2Funderstanding-laboratory-glassware-classifications%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-26T17%3A09%3A58Z%22%7D%7D%2C%7B%22key%22%3A%224ET35UTR%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BLaboratory%20Beakers%20Guide%20%26%23x2013%3B%20Types%20%26amp%3B%20Uses%20Explained%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20ChemScience%20Inc.%20Retrieved%20February%2026%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.chemscience.com%5C%2Fblog%5C%2Fwww.chemscience.com%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.chemscience.com%5C%2Fblog%5C%2Fwww.chemscience.com%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Laboratory%20Beakers%20Guide%20%5Cu2013%20Types%20%26%20Uses%20Explained%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22Explore%20different%20beaker%20types%20glass%2C%20plastic%2C%20metal%20for%20accurate%20lab%20use.%20Find%20tips%20for%20safe%20handling%20and%20choosing%20the%20right%20container%20for%20better%20accuracy%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.chemscience.com%5C%2Fblog%5C%2Fwww.chemscience.com%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-26T17%3A09%3A20Z%22%7D%7D%2C%7B%22key%22%3A%22NK7965IH%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Carolina.Biological%22%2C%22parsedDate%22%3A%222020-01-01%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BCarolina.Biological.%20%282020%2C%20January%201%29.%20%26lt%3Bi%26gt%3BStocking%20a%20laboratory%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fknowledge.carolina.com%5C%2Fproduct-resources%5C%2Fbuying-guides%5C%2Fultimate-lab-basics-buying-guide%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fknowledge.carolina.com%5C%2Fproduct-resources%5C%2Fbuying-guides%5C%2Fultimate-lab-basics-buying-guide%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Stocking%20a%20laboratory%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22Carolina.Biological%22%7D%5D%2C%22abstractNote%22%3A%22Start%20the%20new%20school%20year%20off%20right%20by%20stocking%20up%20on%20top-brand%20supplies%20from%20a%20name%20you%20can%20trust.%22%2C%22blogTitle%22%3A%22%22%2C%22date%22%3A%222020-01-01T20%3A26%3A23-05%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fknowledge.carolina.com%5C%2Fproduct-resources%5C%2Fbuying-guides%5C%2Fultimate-lab-basics-buying-guide%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-26T17%3A02%3A58Z%22%7D%7D%2C%7B%22key%22%3A%2282RD8EEB%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Lam%20et%20al.%22%2C%22parsedDate%22%3A%222024-05-21%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BLam%2C%20K.%2C%20Armstrong%2C%20B.%2C%20%26amp%3B%20Margelefsky%2C%20E.%20L.%20%282024%29.%20Correction%20to%20%26%23x201C%3BGravimetric%20and%20Thermal-Imaging%20Characterization%20of%20Water-Free%20Reflux%20Condensers%20for%20Laboratory%20Applications.%26%23x201D%3B%20%26lt%3Bi%26gt%3BACS%20Omega%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B9%26lt%3B%5C%2Fi%26gt%3B%2820%29%2C%2022508.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1021%5C%2Facsomega.4c03508%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1021%5C%2Facsomega.4c03508%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Correction%20to%20%5C%22Gravimetric%20and%20Thermal-Imaging%20Characterization%20of%20Water-Free%20Reflux%20Condensers%20for%20Laboratory%20Applications%5C%22%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kevin%22%2C%22lastName%22%3A%22Lam%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Brittany%22%2C%22lastName%22%3A%22Armstrong%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eric%20L.%22%2C%22lastName%22%3A%22Margelefsky%22%7D%5D%2C%22abstractNote%22%3A%22%5BThis%20corrects%20the%20article%20DOI%3A%2010.1021%5C%2Facsomega.4c00411.%5D.%22%2C%22date%22%3A%222024-05-21%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1021%5C%2Facsomega.4c03508%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222470-1343%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-18T20%3A03%3A14Z%22%7D%7D%2C%7B%22key%22%3A%22F4YXNT8D%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Lam%20et%20al.%22%2C%22parsedDate%22%3A%222024-04-09%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BLam%2C%20K.%2C%20Armstrong%2C%20B.%2C%20%26amp%3B%20Margelefsky%2C%20E.%20L.%20%282024%29.%20Gravimetric%20and%20Thermal-Imaging%20Characterization%20of%20Water-Free%20Reflux%20Condensers%20for%20Laboratory%20Applications.%20%26lt%3Bi%26gt%3BACS%20Omega%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B9%26lt%3B%5C%2Fi%26gt%3B%2814%29%2C%2016563%26%23x2013%3B16571.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1021%5C%2Facsomega.4c00411%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1021%5C%2Facsomega.4c00411%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Gravimetric%20and%20Thermal-Imaging%20Characterization%20of%20Water-Free%20Reflux%20Condensers%20for%20Laboratory%20Applications%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kevin%22%2C%22lastName%22%3A%22Lam%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Brittany%22%2C%22lastName%22%3A%22Armstrong%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eric%20L.%22%2C%22lastName%22%3A%22Margelefsky%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222024-04-09%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1021%5C%2Facsomega.4c00411%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fpubs.acs.org%5C%2Fdoi%5C%2F10.1021%5C%2Facsomega.4c00411%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222470-1343%2C%202470-1343%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-18T20%3A02%3A52Z%22%7D%7D%2C%7B%22key%22%3A%228G9YUDWQ%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Fordham%22%2C%22parsedDate%22%3A%222024-02-07%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BFordham%2C%20C.%20%282024%2C%20February%207%29.%20Condenser%20Chemistry%3A%20What%26%23x2019%3Bs%20the%20Difference%20Between%20Distillation%20%26amp%3B%20Reflux%3F%20%26lt%3Bi%26gt%3BAsynt%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.asynt.com%5C%2Fblog%5C%2Fcondenser-chemistry-difference-between-distillation-reflux%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.asynt.com%5C%2Fblog%5C%2Fcondenser-chemistry-difference-between-distillation-reflux%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Condenser%20Chemistry%3A%20What%27s%20the%20Difference%20Between%20Distillation%20%26%20Reflux%3F%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cameron%22%2C%22lastName%22%3A%22Fordham%22%7D%5D%2C%22abstractNote%22%3A%22Explore%20the%20differences%20between%20distillation%20and%20reflux%20in%20condenser%20chemistry%2C%20from%20simple%20to%20vacuum%20distillation%20and%20their%20key%20applications.%22%2C%22blogTitle%22%3A%22Asynt%22%2C%22date%22%3A%222024-02-07T12%3A41%3A42%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.asynt.com%5C%2Fblog%5C%2Fcondenser-chemistry-difference-between-distillation-reflux%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-GB%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-18T19%3A36%3A52Z%22%7D%7D%2C%7B%22key%22%3A%227QMSRV59%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Fordham%22%2C%22parsedDate%22%3A%222024-02-07%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BFordham%2C%20C.%20%282024%2C%20February%207%29.%20Condenser%20Chemistry%3A%20What%26%23x2019%3Bs%20the%20Difference%20Between%20Distillation%20%26amp%3B%20Reflux%3F%20%26lt%3Bi%26gt%3BAsynt%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.asynt.com%5C%2Fblog%5C%2Fcondenser-chemistry-difference-between-distillation-reflux%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.asynt.com%5C%2Fblog%5C%2Fcondenser-chemistry-difference-between-distillation-reflux%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Condenser%20Chemistry%3A%20What%27s%20the%20Difference%20Between%20Distillation%20%26%20Reflux%3F%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cameron%22%2C%22lastName%22%3A%22Fordham%22%7D%5D%2C%22abstractNote%22%3A%22Explore%20the%20differences%20between%20distillation%20and%20reflux%20in%20condenser%20chemistry%2C%20from%20simple%20to%20vacuum%20distillation%20and%20their%20key%20applications.%22%2C%22blogTitle%22%3A%22Asynt%22%2C%22date%22%3A%222024-02-07T12%3A41%3A42%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.asynt.com%5C%2Fblog%5C%2Fcondenser-chemistry-difference-between-distillation-reflux%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-GB%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-18T19%3A23%3A25Z%22%7D%7D%2C%7B%22key%22%3A%229QSHPRKP%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22admin%22%2C%22parsedDate%22%3A%222025-10-28%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3Badmin.%20%282025%2C%20October%2028%29.%20What%20Is%20Reflux%20Distillation%3F%20A%20Complete%20Guide.%20%26lt%3Bi%26gt%3BBorosil%20Scientific%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.borosilscientific.com%5C%2Fwhat-is-reflux-distillation-complete-guide%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.borosilscientific.com%5C%2Fwhat-is-reflux-distillation-complete-guide%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22What%20Is%20Reflux%20Distillation%3F%20A%20Complete%20Guide%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22admin%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22blogTitle%22%3A%22Borosil%20Scientific%22%2C%22date%22%3A%222025-10-28T12%3A03%3A55%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.borosilscientific.com%5C%2Fwhat-is-reflux-distillation-complete-guide%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-18T19%3A22%3A37Z%22%7D%7D%2C%7B%22key%22%3A%22M3IV7IHG%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Reflux%20and%20distillation%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BReflux%20and%20distillation.%20%28n.d.%29.%20%26lt%3Bi%26gt%3BReflux%20and%20distillation%20%26%23x2013%3B%20practical%20videos%20%7C%2016%26%23x2013%3B18%20students%26lt%3B%5C%2Fi%26gt%3B.%20RSC%20Education.%20Retrieved%20February%2018%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fedu.rsc.org%5C%2Fpractical%5C%2Freflux-and-distillation-practical-videos-16-18-students%5C%2F4012294.article%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fedu.rsc.org%5C%2Fpractical%5C%2Freflux-and-distillation-practical-videos-16-18-students%5C%2F4012294.article%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Reflux%20and%20distillation%20%5Cu2013%20practical%20videos%20%7C%2016%5Cu201318%20students%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22Reflux%20and%20distillation%22%7D%5D%2C%22abstractNote%22%3A%22Learn%20how%20to%20carry%20out%20two%20common%20practical%20techniques%2C%20understand%20how%20they%20differ%20and%20their%20applications.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fedu.rsc.org%5C%2Fpractical%5C%2Freflux-and-distillation-practical-videos-16-18-students%5C%2F4012294.article%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-18T19%3A21%3A05Z%22%7D%7D%2C%7B%22key%22%3A%22BTDJWD9C%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BWater%20flow%20in%20condenser%20-%20ECHEMI.com%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20ECHEMI.%20Retrieved%20February%2018%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.echemi.com%5C%2Fcommunity%5C%2Fwater-flow-in-condenser_mjart2205112471_431.html%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.echemi.com%5C%2Fcommunity%5C%2Fwater-flow-in-condenser_mjart2205112471_431.html%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Water%20flow%20in%20condenser%20-%20ECHEMI.com%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22How%20should%20the%20water%20flow%20in%20your%20condenser%3F%20From%20the%20flask%20to%20the%20top%20of%20the%20condenser%2C%20or%20from%20the%20top%20of%20the%20condenser%20back%20to%20the%20flask%3F%20I%20could%20imagine%20both%3A%20flask%20to%20top%20will%20fill%20the%20entire%20condenser%20with%20water%2C%20because%20of%20the%20resistance%20of%20gravity%2C%20enlarging%20cooling%20area%2C%20or%20top%20to%20flask%2C%20because%20this%20will%20generate%20counter%20current%20exchange%20increasing%20heat%20flow%2C%20but%20then%20it%20is%20almost%20impossible%20to%20get%20a%20decent%20cooling%20surface%20without%20extreme%20water%20pressure.This%20picture%20suggests%20the%20first.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.echemi.com%5C%2Fcommunity%5C%2Fwater-flow-in-condenser_mjart2205112471_431.html%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-18T18%3A22%3A45Z%22%7D%7D%2C%7B%22key%22%3A%22M5PCE2WR%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Abrahams%20and%20Millar%22%2C%22parsedDate%22%3A%222008-11-17%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BAbrahams%2C%20I.%2C%20%26amp%3B%20Millar%2C%20R.%20%282008%29.%20Does%20Practical%20Work%20Really%20Work%3F%20A%20study%20of%20the%20effectiveness%20of%20practical%20work%20as%20a%20teaching%20and%20learning%20method%20in%20school%20science.%20%26lt%3Bi%26gt%3BInternational%20Journal%20of%20Science%20Education%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B30%26lt%3B%5C%2Fi%26gt%3B%2814%29%2C%201945%26%23x2013%3B1969.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1080%5C%2F09500690701749305%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1080%5C%2F09500690701749305%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Does%20Practical%20Work%20Really%20Work%3F%20A%20study%20of%20the%20effectiveness%20of%20practical%20work%20as%20a%20teaching%20and%20learning%20method%20in%20school%20science%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ian%22%2C%22lastName%22%3A%22Abrahams%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Robin%22%2C%22lastName%22%3A%22Millar%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222008-11-17%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1080%5C%2F09500690701749305%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.tandfonline.com%5C%2Fdoi%5C%2Fabs%5C%2F10.1080%5C%2F09500690701749305%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220950-0693%2C%201464-5289%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-18T18%3A05%3A56Z%22%7D%7D%2C%7B%22key%22%3A%22IIMT5KZS%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Vogel%22%2C%22parsedDate%22%3A%221977%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVogel%2C%20A.%20I.%20%281977%29.%20%26lt%3Bi%26gt%3BA%20text-book%20of%20practical%20organic%20chemistry%3A%20incl.%20qualitative%20organic%20analysis%26lt%3B%5C%2Fi%26gt%3B%20%283.%20ed.%3B%20new%20impr%29.%20Longmans.%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22book%22%2C%22title%22%3A%22A%20text-book%20of%20practical%20organic%20chemistry%3A%20incl.%20qualitative%20organic%20analysis%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Arthur%20Israel%22%2C%22lastName%22%3A%22Vogel%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%221977%22%2C%22originalDate%22%3A%22%22%2C%22originalPublisher%22%3A%22%22%2C%22originalPlace%22%3A%22%22%2C%22format%22%3A%22%22%2C%22ISBN%22%3A%229780582442450%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-18T17%3A48%3A38Z%22%7D%7D%2C%7B%22key%22%3A%22YECMMI5M%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Indiana%22%2C%22parsedDate%22%3A%222025-10-22%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BIndiana%2C%20W.%20Q.%20in.%20%282025%2C%20October%2022%29.%20%26lt%3Bi%26gt%3BCommunity%20Drinking%20Water%20Information%20-%20Madison%20County%26lt%3B%5C%2Fi%26gt%3B.%20Water%20Quality%20in%20Indiana.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.in.gov%5C%2Fidem%5C%2Fcleanwater%5C%2Fdrinking-water%5C%2Fcommunity-drinking-water-info-madison-co%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.in.gov%5C%2Fidem%5C%2Fcleanwater%5C%2Fdrinking-water%5C%2Fcommunity-drinking-water-info-madison-co%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Community%20Drinking%20Water%20Information%20-%20Madison%20County%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Water%20Quality%20in%22%2C%22lastName%22%3A%22Indiana%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222025-10-22T15%3A53%3A10-04%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.in.gov%5C%2Fidem%5C%2Fcleanwater%5C%2Fdrinking-water%5C%2Fcommunity-drinking-water-info-madison-co%5C%2F%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-06T21%3A40%3A59Z%22%7D%7D%2C%7B%22key%22%3A%224NDMQFUB%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Schneider%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BSchneider%2C%20K.%20%28n.d.%29.%20%26lt%3Bi%26gt%3BThese%20Indiana%20residents%20have%20brown%20water%20and%20want%20their%20town%20to%20tell%20them%20why%26lt%3B%5C%2Fi%26gt%3B.%20The%20Indianapolis%20Star.%20Retrieved%20February%205%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.indystar.com%5C%2Fstory%5C%2Fnews%5C%2Fenvironment%5C%2F2025%5C%2F10%5C%2F06%5C%2Findiana-brown-water-quality-safe-drinking%5C%2F86481832007%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.indystar.com%5C%2Fstory%5C%2Fnews%5C%2Fenvironment%5C%2F2025%5C%2F10%5C%2F06%5C%2Findiana-brown-water-quality-safe-drinking%5C%2F86481832007%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22These%20Indiana%20residents%20have%20brown%20water%20and%20want%20their%20town%20to%20tell%20them%20why%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Karl%22%2C%22lastName%22%3A%22Schneider%22%7D%5D%2C%22abstractNote%22%3A%22Residents%20in%20Lapel%2C%20Indiana%2C%20are%20experiencing%20brown%20water%20coming%20out%20of%20their%20taps%20and%20are%20worried%20that%20the%20water%20is%20not%20safe%20to%20drink.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.indystar.com%5C%2Fstory%5C%2Fnews%5C%2Fenvironment%5C%2F2025%5C%2F10%5C%2F06%5C%2Findiana-brown-water-quality-safe-drinking%5C%2F86481832007%5C%2F%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T20%3A38%3A48Z%22%7D%7D%2C%7B%22key%22%3A%2266VVKVCR%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BFixed%20Costs%20vs%20Variable%20Costs%3A%20A%20Practical%20Guide%20with%20Examples%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20The%20Fino%20Partners.%20Retrieved%20February%205%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fthefinopartners.com%5C%2Fblogs%5C%2Ffixed-costs-vs-variable-costs-a-practical-guide-with-examples%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fthefinopartners.com%5C%2Fblogs%5C%2Ffixed-costs-vs-variable-costs-a-practical-guide-with-examples%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Fixed%20Costs%20vs%20Variable%20Costs%3A%20A%20Practical%20Guide%20with%20Examples%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22A%20fixed%20cost%20is%20any%20business%20expense%20that%20doesn%26%23039%3Bt%20change%20with%20the%20number%20of%20units%20your%20business%20produces%2C%20sales%20it%20makes%2C%20or%20revenue%20it%20generates.%20A%20fixed%20cost%20is%20a%20firm%20expense%20you%20can%20predict%20every%20single%20time.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fthefinopartners.com%5C%2Fblogs%5C%2Ffixed-costs-vs-variable-costs-a-practical-guide-with-examples%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T16%3A16%3A05Z%22%7D%7D%2C%7B%22key%22%3A%22PJT2KBBP%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22wtolson1%22%2C%22parsedDate%22%3A%222025-11-03%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3Bwtolson1.%20%282025%2C%20November%203%29.%20R%26amp%3BD%20Tax%20Credits%20and%20Deductions%20Explained.%20%26lt%3Bi%26gt%3BBloomberg%20Tax%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fpro.bloombergtax.com%5C%2Finsights%5C%2Ffederal-tax%5C%2Frd-tax-credit-and-deducting-rd-expenditures%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fpro.bloombergtax.com%5C%2Finsights%5C%2Ffederal-tax%5C%2Frd-tax-credit-and-deducting-rd-expenditures%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22R%26D%20Tax%20Credits%20and%20Deductions%20Explained%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22wtolson1%22%7D%5D%2C%22abstractNote%22%3A%22R%26amp%3BD%20tax%20credits%20can%20save%20your%20organization%20thousands%20of%20dollars%20per%20year.%20Learn%20more%20about%20these%20credits%20and%20deductions%2C%20if%20you%20qualify%2C%20and%20more.%22%2C%22blogTitle%22%3A%22Bloomberg%20Tax%22%2C%22date%22%3A%222025-11-03T13%3A03%3A57%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fpro.bloombergtax.com%5C%2Finsights%5C%2Ffederal-tax%5C%2Frd-tax-credit-and-deducting-rd-expenditures%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A47%3A39Z%22%7D%7D%2C%7B%22key%22%3A%22NDQW9KSQ%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Ohnstad%22%2C%22parsedDate%22%3A%222024-07-31%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BOhnstad%2C%20E.%20%282024%2C%20July%2031%29.%20Can%20your%20business%20qualify%20for%20the%20R%26amp%3BD%20tax%20credit%3F%20%26lt%3Bi%26gt%3BAbdo%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fabdosolutions.com%5C%2Fcan-your-business-qualify-for-the-rd-tax-credit%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fabdosolutions.com%5C%2Fcan-your-business-qualify-for-the-rd-tax-credit%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Can%20your%20business%20qualify%20for%20the%20R%26D%20tax%20credit%3F%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elly%22%2C%22lastName%22%3A%22Ohnstad%22%7D%5D%2C%22abstractNote%22%3A%22Unlock%20the%20potential%20of%20Research%20%26amp%3B%20Development%20Tax%20Credits%20for%20your%20industry.%20Explore%20how%20businesses%20in%20manufacturing%2C%20building%20%26amp%3B%20remodeling%2C%20software%2C%20engineering%2C%20and%20more%20can%20qualify%20for%20R%26amp%3BD%20tax%20credits.%22%2C%22blogTitle%22%3A%22Abdo%22%2C%22date%22%3A%222024-07-31T18%3A30%3A32%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fabdosolutions.com%5C%2Fcan-your-business-qualify-for-the-rd-tax-credit%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A47%3A35Z%22%7D%7D%2C%7B%22key%22%3A%2225FX7ZRB%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Gibson%22%2C%22parsedDate%22%3A%222023-03-03%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BGibson%2C%20T.%20%282023%2C%20March%203%29.%205%20Advantages%20of%20the%20Small%20Business%20R%26amp%3BD%20Tax%20Credit.%20%26lt%3Bi%26gt%3BOmega%20Accounting%20Solutions%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fomega-accounting.com%5C%2Fsmall-business-r-d-tax-credit%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fomega-accounting.com%5C%2Fsmall-business-r-d-tax-credit%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%225%20Advantages%20of%20the%20Small%20Business%20R%26D%20Tax%20Credit%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Timothy%22%2C%22lastName%22%3A%22Gibson%22%7D%5D%2C%22abstractNote%22%3A%22R%26amp%3BD%20tax%20credits%20are%20for%20companies%20of%20all%20sizes%2C%20and%20you%20don%5Cu2019t%20have%20to%20cure%20cancer%20to%20qualify.%20Learn%20why%20small%20businesses%20may%20be%20eligible.%22%2C%22blogTitle%22%3A%22Omega%20Accounting%20Solutions%22%2C%22date%22%3A%222023-03-03T00%3A00%3A00%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fomega-accounting.com%5C%2Fsmall-business-r-d-tax-credit%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A47%3A31Z%22%7D%7D%2C%7B%22key%22%3A%2275XH59FV%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Council%22%2C%22parsedDate%22%3A%222025-08-04%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BCouncil%2C%20S.%20B.%20E.%20%282025%2C%20August%204%29.%20%26%23x201C%3BOne%20Big%20Beautiful%20Bill%26%23x201D%3B%20Encourages%20Small%20Business%20Innovation%3A%20How%20Entrepreneurs%20Can%20Immediately%20Benefit%20from%20Restored%20R%26amp%3BD%20Expensing.%20%26lt%3Bi%26gt%3BSmall%20Business%20%26amp%3B%20Entrepreneurship%20Council%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fsbecouncil.org%5C%2F2025%5C%2F08%5C%2F04%5C%2Fone-big-beautiful-bill-encourages-small-business-innovation-how-entrepreneurs-can-immediately-benefit-from-restored-rd-expensing%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fsbecouncil.org%5C%2F2025%5C%2F08%5C%2F04%5C%2Fone-big-beautiful-bill-encourages-small-business-innovation-how-entrepreneurs-can-immediately-benefit-from-restored-rd-expensing%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22%5Cu201cOne%20Big%20Beautiful%20Bill%5Cu201d%20Encourages%20Small%20Business%20Innovation%3A%20How%20Entrepreneurs%20Can%20Immediately%20Benefit%20from%20Restored%20R%26D%20Expensing%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%20B.%20E.%22%2C%22lastName%22%3A%22Council%22%7D%5D%2C%22abstractNote%22%3A%22By%20Barbara%20Weltman%20%5Cu2013%20You%20don%5Cu2019t%20have%20to%20be%20in%20the%20pharmaceutical%20industry%20or%20develop%20software%20to%20engage%20in%20research%20and%20development%20%28R%26amp%3BD%29%20activities%20that%20can%20produce%20new%20things%20for%20sale%20or%20create%20new%20processes%20for%20your%20internal%20business%20operations.%20If%20you%20spend%20money%20on%20R%26amp%3BD%2C%20you%20can%20recoup%20some%20of%20your%20costs%20through%20federal%20%5B%5Cu2026%5D%22%2C%22blogTitle%22%3A%22Small%20Business%20%26%20Entrepreneurship%20Council%22%2C%22date%22%3A%222025-08-04T19%3A56%3A09%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fsbecouncil.org%5C%2F2025%5C%2F08%5C%2F04%5C%2Fone-big-beautiful-bill-encourages-small-business-innovation-how-entrepreneurs-can-immediately-benefit-from-restored-rd-expensing%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A47%3A26Z%22%7D%7D%2C%7B%22key%22%3A%227SP6I8E5%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BWater%20Quality%20Testing%20Business%20Plan%20Report%202026%20%7C%20Setup%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20Retrieved%20February%205%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.imarcgroup.com%5C%2Fwater-quality-testing-business-plan-project-report%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.imarcgroup.com%5C%2Fwater-quality-testing-business-plan-project-report%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Water%20Quality%20Testing%20Business%20Plan%20Report%202026%20%7C%20Setup%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22How%20to%20start%20water%20quality%20testing%20business%3F%20Explore%20buisness%20plan%2C%20industry%20trends%2C%20business%20setup%2C%20revenue%20model%2C%20investment%2C%20expenses%20%26amp%3B%20profitability.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.imarcgroup.com%5C%2Fwater-quality-testing-business-plan-project-report%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A47%3A06Z%22%7D%7D%2C%7B%22key%22%3A%22ELSX67YZ%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Brockovich%22%2C%22parsedDate%22%3A%222025-11-12%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BBrockovich%2C%20E.%20%282025%2C%20November%2012%29.%20%26lt%3Bi%26gt%3BWho%26%23x2019%3Bs%20In%20Charge%20Of%20The%20Water%20In%20Madison%20County%2C%20Indiana%3F%26lt%3B%5C%2Fi%26gt%3B%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.thebrockovichreport.com%5C%2Fp%5C%2Fwhos-in-charge-of-the-water-in-madison%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.thebrockovichreport.com%5C%2Fp%5C%2Fwhos-in-charge-of-the-water-in-madison%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Who%27s%20In%20Charge%20Of%20The%20Water%20In%20Madison%20County%2C%20Indiana%3F%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Erin%22%2C%22lastName%22%3A%22Brockovich%22%7D%5D%2C%22abstractNote%22%3A%22Local%20Officials%20Fumble%20As%20Residents%20Raise%20Concerns%20About%20The%20Look%20Of%20Their%20Water%22%2C%22date%22%3A%222025-11-12%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.thebrockovichreport.com%5C%2Fp%5C%2Fwhos-in-charge-of-the-water-in-madison%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A43%3A45Z%22%7D%7D%2C%7B%22key%22%3A%22BVVF7B89%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Amritha%20R%22%2C%22parsedDate%22%3A%222024-09-22%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BAmritha%20R.%20%282024%2C%20September%2022%29.%20%26lt%3Bi%26gt%3BTHE%20IMPACT%20OF%20VARIABLE%20AND%20FIXED%20COST%20ON%20BUSINESS%20DECISION-MAKING%20%7C%20LinkedIn%26lt%3B%5C%2Fi%26gt%3B%20%5BSocial%20Media%5D.%20Geojit%20Investments%20Limited.%20LinkedIn.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.linkedin.com%5C%2Fpulse%5C%2Fimpact-variable-fixed-cost-business-decision-making-amritha-r-3axbc%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.linkedin.com%5C%2Fpulse%5C%2Fimpact-variable-fixed-cost-business-decision-making-amritha-r-3axbc%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22THE%20IMPACT%20OF%20VARIABLE%20AND%20FIXED%20COST%20ON%20BUSINESS%20DECISION-MAKING%20%7C%20LinkedIn%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22Amritha%20R%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222024-09-22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.linkedin.com%5C%2Fpulse%5C%2Fimpact-variable-fixed-cost-business-decision-making-amritha-r-3axbc%5C%2F%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A35%3A55Z%22%7D%7D%2C%7B%22key%22%3A%22ZD9SIRUP%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22365outsource_admin%22%2C%22parsedDate%22%3A%222025-10-24%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B365outsource_admin.%20%282025%2C%20October%2024%29.%20How%20Small%20Businesses%20Cut%20Costs%20with%20Outsourcing.%20%26lt%3Bi%26gt%3B365Outsource.Com%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.365outsource.com%5C%2Funcategorized%5C%2Fsmall-businesses-cut-costs-outsourcing%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.365outsource.com%5C%2Funcategorized%5C%2Fsmall-businesses-cut-costs-outsourcing%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22How%20Small%20Businesses%20Cut%20Costs%20with%20Outsourcing%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22365outsource_admin%22%7D%5D%2C%22abstractNote%22%3A%22Learn%20how%20small%20businesses%20can%20save%20costs%2C%20access%20expertise%2C%20and%20enhance%20efficiency%20through%20strategic%20outsourcing.%22%2C%22blogTitle%22%3A%22365Outsource.com%22%2C%22date%22%3A%222025-10-24T04%3A19%3A53%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.365outsource.com%5C%2Funcategorized%5C%2Fsmall-businesses-cut-costs-outsourcing%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A18%3A15Z%22%7D%7D%2C%7B%22key%22%3A%22J2K44NWR%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Sullivan%22%2C%22parsedDate%22%3A%222023-09-19%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BSullivan%2C%20P.%20%282023%2C%20September%2019%29.%20Why%20You%20Need%20a%20Third-Party%20Assessment%20for%20Cybersecurity.%20%26lt%3Bi%26gt%3BA-LIGN%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.a-lign.com%5C%2Farticles%5C%2Fwhy-you-need-a-third-party-assessment-for-cybersecurity%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.a-lign.com%5C%2Farticles%5C%2Fwhy-you-need-a-third-party-assessment-for-cybersecurity%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Why%20You%20Need%20a%20Third-Party%20Assessment%20for%20Cybersecurity%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Patrick%22%2C%22lastName%22%3A%22Sullivan%22%7D%5D%2C%22abstractNote%22%3A%22While%20a%20self-assessment%20can%20be%20the%20initial%20start%20to%20a%20compliance%20journey%2C%20a%20third-party%20assessment%20is%20not%20just%20preferable%2C%20but%20essential.%22%2C%22blogTitle%22%3A%22A-LIGN%22%2C%22date%22%3A%222023-09-19T12%3A56%3A28%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.a-lign.com%5C%2Farticles%5C%2Fwhy-you-need-a-third-party-assessment-for-cybersecurity%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A12%3A08Z%22%7D%7D%2C%7B%22key%22%3A%22RWBMED99%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BWhat%20Is%20Third-Party%20Risk%20Management%3F%26lt%3B%5C%2Fi%26gt%3B%20%28n.d.%29.%20Retrieved%20February%205%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.venminder.com%5C%2Fblog%5C%2Fwhat-is-third-party-risk-management%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.venminder.com%5C%2Fblog%5C%2Fwhat-is-third-party-risk-management%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22What%20Is%20Third-Party%20Risk%20Management%3F%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22Third-party%20risk%20management%20helps%20identify%2C%20assess%2C%20manage%2C%20%26amp%3B%20monitor%20vendor%20risks%20posed%20to%20your%20organization.%20Learn%20what%20third-party%20risk%20management%20is.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.venminder.com%5C%2Fblog%5C%2Fwhat-is-third-party-risk-management%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A12%3A03Z%22%7D%7D%2C%7B%22key%22%3A%22CGX4UKLZ%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BWater%20Quality%20Studies%20%26%23x2013%3B%20Johnson%20Engineering%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20Retrieved%20February%205%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fjohnsonengineering.com%5C%2Fservices-projects%5C%2Fwater-quality-studies%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fjohnsonengineering.com%5C%2Fservices-projects%5C%2Fwater-quality-studies%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Water%20Quality%20Studies%20%5Cu2013%20Johnson%20Engineering%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22%22%2C%22blogTitle%22%3A%22%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fjohnsonengineering.com%5C%2Fservices-projects%5C%2Fwater-quality-studies%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A09%3A06Z%22%7D%7D%2C%7B%22key%22%3A%22XJQSDMLI%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22parsedDate%22%3A%222025-06-26%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BWater%20Treatment%20Feasibility%20Studies%20-%20AQUASGROUP%26lt%3B%5C%2Fi%26gt%3B.%20%282025%2C%20June%2026%29.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Faquasgroup.com%5C%2Fservice%5C%2Fion-exchange-resin-regeneration-activated-carbon-reactivation-services%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Faquasgroup.com%5C%2Fservice%5C%2Fion-exchange-resin-regeneration-activated-carbon-reactivation-services%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Water%20Treatment%20Feasibility%20Studies%20-%20AQUASGROUP%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22ISO%209001%3A2015%20certified%20ion%20exchange%20resin%20regeneration%20and%20GAC%20reactivation%20services%20for%20industrial%20process%20water%20in%20aerospace%2C%20medical%20device%2C%20electronics%2C%20and%20more.%22%2C%22date%22%3A%222025-06-26T14%3A40%3A38%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Faquasgroup.com%5C%2Fservice%5C%2Fion-exchange-resin-regeneration-activated-carbon-reactivation-services%5C%2F%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A09%3A02Z%22%7D%7D%2C%7B%22key%22%3A%22GTUH53EI%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BLife%20Science%20Content%20Marketing%20Services.%20%28n.d.%29.%20%26lt%3Bi%26gt%3BSciencia%20Consulting%26lt%3B%5C%2Fi%26gt%3B.%20Retrieved%20February%205%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fscienciaconsulting.com%5C%2Fcontent-marketing-for-life-sciences%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fscienciaconsulting.com%5C%2Fcontent-marketing-for-life-sciences%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Life%20Science%20Content%20Marketing%20Services%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22Boost%20your%20brand%20with%20expert%20life%20science%20content%20marketing.%20Sciencia%20Consulting%20delivers%20compliant%2C%20engaging%20content%20to%20drive%20traffic%20and%20trust.%22%2C%22blogTitle%22%3A%22Sciencia%20Consulting%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fscienciaconsulting.com%5C%2Fcontent-marketing-for-life-sciences%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T15%3A08%3A55Z%22%7D%7D%2C%7B%22key%22%3A%22VYKM956X%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BNew%20Evidence%20That%20the%20R%26amp%3BD%20Tax%20Credit%20Promotes%20Research%2C%20Especially%20in%20Small%20Companies%20%7C%20ITIF%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20Retrieved%20February%205%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fitif.org%5C%2Fpublications%5C%2F2020%5C%2F05%5C%2F05%5C%2Fnew-evidence-rd-tax-credit-promotes-research-especially-small-companies%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fitif.org%5C%2Fpublications%5C%2F2020%5C%2F05%5C%2F05%5C%2Fnew-evidence-rd-tax-credit-promotes-research-especially-small-companies%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22New%20Evidence%20That%20the%20R%26D%20Tax%20Credit%20Promotes%20Research%2C%20Especially%20in%20Small%20Companies%20%7C%20ITIF%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fitif.org%5C%2Fpublications%5C%2F2020%5C%2F05%5C%2F05%5C%2Fnew-evidence-rd-tax-credit-promotes-research-especially-small-companies%5C%2F%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T14%3A44%3A54Z%22%7D%7D%2C%7B%22key%22%3A%22DCTFA8QU%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22parsedDate%22%3A%222016-03-08%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BThe%20Type%20of%20R%26amp%3BD%20That%20Small%20Businesses%20Can%20Really%20Use%20-%20NicheLabs%2C%20LLC.%20%282016%2C%20March%208%29.%20%26lt%3Bi%26gt%3BNicheLabs%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.nichelabs.com%5C%2Fgeneral%5C%2Fthe-type-of-rd-that-small-businesses-can-really-use%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.nichelabs.com%5C%2Fgeneral%5C%2Fthe-type-of-rd-that-small-businesses-can-really-use%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22The%20Type%20of%20R%26D%20That%20Small%20Businesses%20Can%20Really%20Use%20-%20NicheLabs%2C%20LLC%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22If%20you%20are%20looking%20to%20grow%20your%20business%20to%20find%20more%20clients%2C%20customers%20or%20patients%20without%20spending%20a%20fortune%2C%20then%20the%20tactic%20that%20my%20friend%20Steve%20Clements%22%2C%22blogTitle%22%3A%22NicheLabs%22%2C%22date%22%3A%222016-03-08T10%3A00%3A58-04%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.nichelabs.com%5C%2Fgeneral%5C%2Fthe-type-of-rd-that-small-businesses-can-really-use%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T14%3A44%3A43Z%22%7D%7D%2C%7B%22key%22%3A%22PUB8TJ97%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22parsedDate%22%3A%222016-03-08%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BThe%20Type%20of%20R%26amp%3BD%20That%20Small%20Businesses%20Can%20Really%20Use%20-%20NicheLabs%2C%20LLC.%20%282016%2C%20March%208%29.%20%26lt%3Bi%26gt%3BNicheLabs%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.nichelabs.com%5C%2Fgeneral%5C%2Fthe-type-of-rd-that-small-businesses-can-really-use%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.nichelabs.com%5C%2Fgeneral%5C%2Fthe-type-of-rd-that-small-businesses-can-really-use%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22The%20Type%20of%20R%26D%20That%20Small%20Businesses%20Can%20Really%20Use%20-%20NicheLabs%2C%20LLC%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22If%20you%20are%20looking%20to%20grow%20your%20business%20to%20find%20more%20clients%2C%20customers%20or%20patients%20without%20spending%20a%20fortune%2C%20then%20the%20tactic%20that%20my%20friend%20Steve%20Clements%22%2C%22blogTitle%22%3A%22NicheLabs%22%2C%22date%22%3A%222016-03-08T10%3A00%3A58-04%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.nichelabs.com%5C%2Fgeneral%5C%2Fthe-type-of-rd-that-small-businesses-can-really-use%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T14%3A44%3A34Z%22%7D%7D%2C%7B%22key%22%3A%22DKN7V5GZ%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22editor%20et%20al.%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3Beditor%2C%20P.%20R.%20F.%20B.%20P.%20R.%20is%20a%20copy%2C%20Economics%2C%20F.-C.%20with%20E.%20in%2C%20Finance%2C%20P.%2C%20%26amp%3B%20policies%2C%20%20over%20twenty%20years%20of%20experience%20in%20the%20classroom%20L.%20about%20our%20editorial.%20%28n.d.%29.%20%26lt%3Bi%26gt%3BHow%20Outsourcing%20Reduces%20Business%20Costs%3A%20Strategies%20and%20Examples%26lt%3B%5C%2Fi%26gt%3B.%20Investopedia.%20Retrieved%20February%205%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.investopedia.com%5C%2Fterms%5C%2Fo%5C%2Foutsourcing.asp%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.investopedia.com%5C%2Fterms%5C%2Fo%5C%2Foutsourcing.asp%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22How%20Outsourcing%20Reduces%20Business%20Costs%3A%20Strategies%20and%20Examples%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pete%20Rathburn%20Full%20Bio%20Pete%20Rathburn%20is%20a%20copy%22%2C%22lastName%22%3A%22editor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fact-Checker%20with%20Expertise%20in%22%2C%22lastName%22%3A%22Economics%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Personal%22%2C%22lastName%22%3A%22Finance%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22over%20twenty%20years%20of%20experience%20in%20the%20classroom%20Learn%20about%20our%20editorial%22%2C%22lastName%22%3A%22policies%22%7D%5D%2C%22abstractNote%22%3A%22Discover%20how%20outsourcing%20can%20lower%20business%20costs%20and%20enhance%20efficiency.%20Learn%20strategies%20with%20practical%20examples%2C%20benefits%2C%20and%20potential%20downsides%20of%20outsourcing.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.investopedia.com%5C%2Fterms%5C%2Fo%5C%2Foutsourcing.asp%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T14%3A41%3A39Z%22%7D%7D%2C%7B%22key%22%3A%22PWPNYP7W%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22365outsource_admin%22%2C%22parsedDate%22%3A%222025-10-24%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B365outsource_admin.%20%282025%2C%20October%2024%29.%20How%20Small%20Businesses%20Cut%20Costs%20with%20Outsourcing.%20%26lt%3Bi%26gt%3B365Outsource.Com%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.365outsource.com%5C%2Funcategorized%5C%2Fsmall-businesses-cut-costs-outsourcing%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.365outsource.com%5C%2Funcategorized%5C%2Fsmall-businesses-cut-costs-outsourcing%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22How%20Small%20Businesses%20Cut%20Costs%20with%20Outsourcing%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22365outsource_admin%22%7D%5D%2C%22abstractNote%22%3A%22Learn%20how%20small%20businesses%20can%20save%20costs%2C%20access%20expertise%2C%20and%20enhance%20efficiency%20through%20strategic%20outsourcing.%22%2C%22blogTitle%22%3A%22365Outsource.com%22%2C%22date%22%3A%222025-10-24T04%3A19%3A53%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.365outsource.com%5C%2Funcategorized%5C%2Fsmall-businesses-cut-costs-outsourcing%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T14%3A41%3A35Z%22%7D%7D%2C%7B%22key%22%3A%22SDTMIP2I%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BThe%20Hidden%20Costs%20of%20R%26amp%3BD%20Outsourcing%20and%20How%20to%20Mitigate%20Them%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20Retrieved%20February%205%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.accelerance.com%5C%2Fblog%5C%2Fthe-hidden-costs-of-outsourcing-rd-and-how-to-mitigate-them%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.accelerance.com%5C%2Fblog%5C%2Fthe-hidden-costs-of-outsourcing-rd-and-how-to-mitigate-them%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22The%20Hidden%20Costs%20of%20R%26D%20Outsourcing%20and%20How%20to%20Mitigate%20Them%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22R%26amp%3BD%20outsourcing%20is%20a%20valid%20method%20for%20reducing%20costs%20and%20increasing%20profits%2C%20but%20organizations%20should%20approach%20the%20practice%20with%20some%20care.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.accelerance.com%5C%2Fblog%5C%2Fthe-hidden-costs-of-outsourcing-rd-and-how-to-mitigate-them%22%2C%22language%22%3A%22en-us%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-02-05T14%3A41%3A27Z%22%7D%7D%2C%7B%22key%22%3A%22JKUHC97D%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Bob%20Kaiser%22%2C%22parsedDate%22%3A%222015-12-23%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BBob%20Kaiser.%20%282015%2C%20December%2023%29.%20Simple%20machines%20%5BEdu%5D.%20%26lt%3Bi%26gt%3BKaiserScience%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fkaiserscience.wordpress.com%5C%2Fphysics%5C%2Fsimple-machines%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fkaiserscience.wordpress.com%5C%2Fphysics%5C%2Fsimple-machines%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Simple%20machines%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22Bob%20Kaiser%22%7D%5D%2C%22abstractNote%22%3A%22Engineering%20is%20the%20use%20of%20physics%20to%20design%20something%20%5Cu2013%20such%20as%20machines%2C%20electrical%20devices%2C%20buildings%2C%20vehicles%2C%20and%20infrastructure.%20In%20this%20lesson%20we%20start%20by%20looking%20at%20the%20most%20basic%20elements%20%5Cu2026%22%2C%22blogTitle%22%3A%22KaiserScience%22%2C%22date%22%3A%222015-12-23T01%3A56%3A32%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fkaiserscience.wordpress.com%5C%2Fphysics%5C%2Fsimple-machines%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-29T14%3A52%3A26Z%22%7D%7D%2C%7B%22key%22%3A%22LSE8GSL3%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22shehlit%22%2C%22parsedDate%22%3A%222021-07-19%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3Bshehlit.%20%282021%2C%20July%2019%29.%20Student%20misconceptions%20about%20work%20and%20energy.%20%26lt%3Bi%26gt%3BChang%20Physics%20Class%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fchangphysicsclass.com%5C%2F2021%5C%2F07%5C%2F19%5C%2Fstudent-misconceptions-about-work-and-energy%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fchangphysicsclass.com%5C%2F2021%5C%2F07%5C%2F19%5C%2Fstudent-misconceptions-about-work-and-energy%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Student%20misconceptions%20about%20work%20and%20energy%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22shehlit%22%7D%5D%2C%22abstractNote%22%3A%22Continuing%20the%20series%20of%20student%20misconceptions%20that%20have%20been%20reported%20by%20the%20physics%20education%20research%20community%2C%20this%20post%20is%20about%20work%20and%20energy.%20Energy%20is%20an%20abstract%20concept.%20It%20cannot%20be%20%5Cu2026%22%2C%22blogTitle%22%3A%22Chang%20Physics%20Class%22%2C%22date%22%3A%222021-07-19T07%3A30%3A00%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fchangphysicsclass.com%5C%2F2021%5C%2F07%5C%2F19%5C%2Fstudent-misconceptions-about-work-and-energy%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-29T14%3A45%3A47Z%22%7D%7D%2C%7B%22key%22%3A%22I9VSTK7C%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BCan%20you%20have%20%26%23x201C%3Btoo%20much%26%23x201D%3B%20mechanical%20advantage%3F%26lt%3B%5C%2Fi%26gt%3B%20%28n.d.%29.%20Alpinesavvy.%20Retrieved%20January%2029%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.alpinesavvy.com%5C%2Fblog%5C%2Fcan-you-have-too-much-mechanical-advantage%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.alpinesavvy.com%5C%2Fblog%5C%2Fcan-you-have-too-much-mechanical-advantage%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Can%20you%20have%20%5C%22too%20much%5C%22%20mechanical%20advantage%3F%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22A%20good%20general%20rule%3A%20use%20the%20least%20amount%20of%20mechanical%20advantage%20%28MA%29%20that%20you%20can%20to%20get%20the%20job%20done.%20Adding%20additional%20pulleys%20and%20ropes%20grabs%20also%20introduces%20more%20friction.%20In%20some%20cases%2C%20what%20you%20think%20is%20giving%20you%20extra%20MA%20may%20actually%20be%20worse%20than%20what%20you%20had%20before.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.alpinesavvy.com%5C%2Fblog%5C%2Fcan-you-have-too-much-mechanical-advantage%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-29T14%3A41%3A41Z%22%7D%7D%2C%7B%22key%22%3A%22DBTVX9QD%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Piatt%22%2C%22parsedDate%22%3A%222025-10-21%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BPiatt%2C%20L.%20%282025%2C%20October%2021%29.%20Understanding%20Pulley%20Systems%20for%20Mechanical%20Advantage.%20%26lt%3Bi%26gt%3BRigging%20Lab%20Academy%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Frigginglabacademy.com%5C%2Fpulley-systems-for-mechanical-advantage%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Frigginglabacademy.com%5C%2Fpulley-systems-for-mechanical-advantage%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Understanding%20Pulley%20Systems%20for%20Mechanical%20Advantage%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lance%22%2C%22lastName%22%3A%22Piatt%22%7D%5D%2C%22abstractNote%22%3A%22Understanding%20pulley%20systems%20for%20mechanical%20advantage%20explains%20the%20five%20core%20rules%20of%20rope%20rigging%2C%20from%20anchored%20systems%20to%20compounding%20setups.%22%2C%22blogTitle%22%3A%22Rigging%20Lab%20Academy%22%2C%22date%22%3A%222025-10-21T05%3A24%3A17%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Frigginglabacademy.com%5C%2Fpulley-systems-for-mechanical-advantage%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-29T13%3A43%3A04Z%22%7D%7D%2C%7B%22key%22%3A%2266XI6LLD%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kleynhans%22%2C%22parsedDate%22%3A%222021%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BKleynhans%2C%20L.%20%282021%29.%20%26lt%3Bi%26gt%3BUnit%203%3A%20Mechanical%20advantage%20of%20simple%20machines%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttp%3A%5C%2F%5C%2Fncvs2.books.nba.co.za%5C%2Fchapter%5C%2Funit-3-mechanical-advantage-of-simple-machines%5C%2F%26%23039%3B%26gt%3Bhttp%3A%5C%2F%5C%2Fncvs2.books.nba.co.za%5C%2Fchapter%5C%2Funit-3-mechanical-advantage-of-simple-machines%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Unit%203%3A%20Mechanical%20advantage%20of%20simple%20machines%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Leigh%22%2C%22lastName%22%3A%22Kleynhans%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222021%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fncvs2.books.nba.co.za%5C%2Fchapter%5C%2Funit-3-mechanical-advantage-of-simple-machines%5C%2F%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-28T19%3A59%3A37Z%22%7D%7D%2C%7B%22key%22%3A%22TBZ48V8P%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Nave%22%2C%22parsedDate%22%3A%222017%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BNave%2C%20C.%20R.%20%282017%29.%20%26lt%3Bi%26gt%3BSimple%20Machines%26lt%3B%5C%2Fi%26gt%3B%20%5BEducational%5D.%20Georgia%20State%20University.%20HyperPhsyics.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttp%3A%5C%2F%5C%2Fhyperphysics.phy-astr.gsu.edu%5C%2Fhbase%5C%2FMechanics%5C%2Fsimmac.html%23c1%26%23039%3B%26gt%3Bhttp%3A%5C%2F%5C%2Fhyperphysics.phy-astr.gsu.edu%5C%2Fhbase%5C%2FMechanics%5C%2Fsimmac.html%23c1%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Simple%20Machines%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Carl%20Rod%22%2C%22lastName%22%3A%22Nave%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222017%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fhyperphysics.phy-astr.gsu.edu%5C%2Fhbase%5C%2FMechanics%5C%2Fsimmac.html%23c1%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-28T19%3A51%3A32Z%22%7D%7D%2C%7B%22key%22%3A%22VT5MM7GG%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kleynhans%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BKleynhans%2C%20L.%20%28n.d.%29.%20%26lt%3Bi%26gt%3BUnit%201%3A%20The%20mechanical%20advantage%20of%20an%20inclined%20plane%26lt%3B%5C%2Fi%26gt%3B.%20Retrieved%20January%2028%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttp%3A%5C%2F%5C%2Fncvs3.books.nba.co.za%5C%2Fchapter%5C%2Funit%25e2%2580%25af1-the-mechanical-advantage-of-an-inclined-plane%5C%2F%26%23039%3B%26gt%3Bhttp%3A%5C%2F%5C%2Fncvs3.books.nba.co.za%5C%2Fchapter%5C%2Funit%25e2%2580%25af1-the-mechanical-advantage-of-an-inclined-plane%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Unit%201%3A%20The%20mechanical%20advantage%20of%20an%20inclined%20plane%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Leigh%22%2C%22lastName%22%3A%22Kleynhans%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%22%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fncvs3.books.nba.co.za%5C%2Fchapter%5C%2Funit%25e2%2580%25af1-the-mechanical-advantage-of-an-inclined-plane%5C%2F%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-28T19%3A34%3A57Z%22%7D%7D%2C%7B%22key%22%3A%22VRGRD9XC%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22parsedDate%22%3A%222017-04-02%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BMechanical%20Advantage%20Explained.%20%282017%2C%20April%202%29.%20%26lt%3Bi%26gt%3BEducated%20Climber.Com%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.educatedclimber.com%5C%2Fmechanical-advantage-explained%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.educatedclimber.com%5C%2Fmechanical-advantage-explained%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22blogPost%22%2C%22title%22%3A%22Mechanical%20Advantage%20Explained%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22Crane%20at%20Prague%20Castle%2C%20photo%20courtesy%20of%20handshouse.org%20%5Cu201cAny%20sufficiently%20advanced%20technology%20is%20indistinguishable%20from%20magic.%5Cu201d%20%5Cu2013%20Arthur%20C.%20Clarke%20%5Cu00a0%20%5Cu201cMechanical%20advantage%20is%20a%20measure%20of%20the%20force%20amplification%20achieved%20by%20using%20a%20tool%2C%20mechanical%20device%20or%20machine%20system.%20Ideally%2C%20the%20device%20preserves%20the%20input%20power%20and%20simply%20trades%20off%20forces%20against%20movement%20to%20%5Cu2026%22%2C%22blogTitle%22%3A%22Educated%20Climber.com%22%2C%22date%22%3A%222017-04-02T10%3A30%3A44%2B00%3A00%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.educatedclimber.com%5C%2Fmechanical-advantage-explained%5C%2F%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en-US%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-28T19%3A25%3A53Z%22%7D%7D%2C%7B%22key%22%3A%228229B8VH%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BSimple%20Machines%3A%20Mechanical%20Advantage%20%7C%20Research%20Starters%20%7C%20EBSCO%20Research%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20EBSCO.%20Retrieved%20January%2028%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.ebsco.com%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.ebsco.com%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Simple%20Machines%3A%20Mechanical%20Advantage%20%7C%20Research%20Starters%20%7C%20EBSCO%20Research%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22%26lt%3Bp%26gt%3BSimple%20machines%20are%20fundamental%20devices%20that%20facilitate%20work%20by%20amplifying%20force%2C%20and%20mechanical%20advantage%20is%20a%20key%20concept%20in%20understanding%20how%20they%20operate.%20Mechanical%20advantage%20occurs%20when%20a%20machine%20increases%20the%20input%20force%20applied%20to%20it%2C%20allowing%20tasks%20to%20be%20accomplished%20more%20easily.%20For%20instance%2C%20a%20lever%2C%20which%20is%20one%20of%20the%20simplest%20machines%2C%20utilizes%20a%20fulcrum%20to%20redirect%20and%20amplify%20force%2C%20depending%20on%20the%20position%20of%20the%20fulcrum.%20Other%20types%20of%20simple%20machines%20include%20the%20wheel%20and%20axle%2C%20pulleys%2C%20inclined%20planes%2C%20wedges%2C%20and%20screws%2C%20each%20with%20unique%20mechanisms%20for%20providing%20mechanical%20advantage.%26lt%3B%5C%2Fp%26gt%3B%5Cn%26lt%3Bp%26gt%3BThe%20efficiency%20of%20a%20simple%20machine%20is%20determined%20by%20comparing%20its%20actual%20mechanical%20advantage%20to%20its%20ideal%20mechanical%20advantage%2C%20which%20represents%20perfect%20performance%20without%20energy%20loss.%20This%20relationship%20underscores%20the%20law%20of%20conservation%20of%20energy%2C%20indicating%20that%20while%20machines%20can%20multiply%20force%2C%20they%20also%20necessitate%20a%20corresponding%20increase%20in%20distance%20moved.%20Consequently%2C%20understanding%20the%20principles%20of%20work%2C%20force%2C%20and%20mechanical%20advantage%20is%20essential%20for%20grasping%20how%20even%20complex%20machinery%20functions%20in%20everyday%20life.%20By%20exploring%20these%20concepts%2C%20individuals%20can%20gain%20insight%20into%20how%20various%20mechanical%20systems%20operate%20and%20improve%20their%20effectiveness%20in%20tasks%20that%20require%20force%20amplification.%26lt%3B%5C%2Fp%26gt%3B%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.ebsco.com%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-28T19%3A00%3A46Z%22%7D%7D%2C%7B%22key%22%3A%22BJC5WG4B%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Urone%20et%20al.%22%2C%22parsedDate%22%3A%222020-03-26%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BUrone%2C%20P.%20P.%2C%20Hinrichs%2C%20R.%2C%20Urone%2C%20P.%20P.%2C%20%26amp%3B%20Hinrichs%2C%20R.%20%282020%2C%20March%2026%29.%20%26lt%3Bi%26gt%3B9.3%20Simple%20Machines%20-%20Physics%20%7C%20OpenStax%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fopenstax.org%5C%2Fbooks%5C%2Fphysics%5C%2Fpages%5C%2F9-3-simple-machines%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fopenstax.org%5C%2Fbooks%5C%2Fphysics%5C%2Fpages%5C%2F9-3-simple-machines%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%229.3%20Simple%20Machines%20-%20Physics%20%7C%20OpenStax%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paul%20Peter%22%2C%22lastName%22%3A%22Urone%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Roger%22%2C%22lastName%22%3A%22Hinrichs%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paul%20Peter%22%2C%22lastName%22%3A%22Urone%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Roger%22%2C%22lastName%22%3A%22Hinrichs%22%7D%5D%2C%22abstractNote%22%3A%22This%20free%20textbook%20is%20an%20OpenStax%20resource%20written%20to%20increase%20student%20access%20to%20high-quality%2C%20peer-reviewed%20learning%20materials.%22%2C%22date%22%3A%22Mar%2026%2C%202020%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fopenstax.org%5C%2Fbooks%5C%2Fphysics%5C%2Fpages%5C%2F9-3-simple-machines%22%2C%22language%22%3A%22English%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-28T18%3A32%3A10Z%22%7D%7D%2C%7B%22key%22%3A%22VIILXYBP%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BSimple%20Machines%20Lesson%20Plan%20%26%23x2013%3B%20A%20Complete%205E%20Method%20Science%20Unit%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20Retrieved%20January%2028%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fkeslerscience.com%5C%2Fsimple-machines-lesson-plan-a-complete-science-lesson-using-the-5e-method-of-instruction%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fkeslerscience.com%5C%2Fsimple-machines-lesson-plan-a-complete-science-lesson-using-the-5e-method-of-instruction%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Simple%20Machines%20Lesson%20Plan%20%5Cu2013%20A%20Complete%205E%20Method%20Science%20Unit%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22Easy%5C%2Fengaging%20Sound%20Waves%205E%20lesson%20plan%20for%20middle%20school%20students%20to%20learn%20about%20and%20identify%20various%20types%20of%20simple%20machines%20and%20how%20they%20change%20force.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fkeslerscience.com%5C%2Fsimple-machines-lesson-plan-a-complete-science-lesson-using-the-5e-method-of-instruction%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-28T17%3A54%3A45Z%22%7D%7D%2C%7B%22key%22%3A%22CPCNRCVR%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3B%26lt%3Bi%26gt%3BSimple%20Machines%3A%20Types%2C%20Activities%2C%20and%20Misconceptions%26lt%3B%5C%2Fi%26gt%3B.%20%28n.d.%29.%20Studylib.Net.%20Retrieved%20January%2028%2C%202026%2C%20from%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fstudylib.net%5C%2Fdoc%5C%2F5444329%5C%2Ffor-forces-and-motion-simple-machines-types-of-simple-mac...%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fstudylib.net%5C%2Fdoc%5C%2F5444329%5C%2Ffor-forces-and-motion-simple-machines-types-of-simple-mac...%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Simple%20Machines%3A%20Types%2C%20Activities%2C%20and%20Misconceptions%22%2C%22creators%22%3A%5B%5D%2C%22abstractNote%22%3A%22Explore%20simple%20machines%20with%20this%20elementary%20science%20presentation.%20Learn%20about%20levers%2C%20inclined%20planes%2C%20wheels%2C%20pulleys%2C%20wedges%2C%20and%20screws.%20Includes%20activity%20ideas.%22%2C%22date%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fstudylib.net%5C%2Fdoc%5C%2F5444329%5C%2Ffor-forces-and-motion-simple-machines-types-of-simple-mac...%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222026-01-28T17%3A53%3A33Z%22%7D%7D%2C%7B%22key%22%3A%2269CNKVTI%22%2C%22library%22%3A%7B%22id%22%3A21495%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Daniel%20J.%20Peppe%20and%20Alan%20L.%20Deino%22%2C%22parsedDate%22%3A%222013%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BDaniel%20J.%20Peppe%2C%20%26amp%3B%20Alan%20L.%20Deino.%20%282013%29.%20%26lt%3Bi%26gt%3BDating%20Rocks%20and%20Fossils%20Using%20Geologic%20Methods%26lt%3B%5C%2Fi%26gt%3B.%20Nature%2C%20Scitable.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.nature.com%5C%2Fscitable%5C%2Fknowledge%5C%2Flibrary%5C%2Fdating-rocks-and-fossils-using-geologic-methods-107924044%5C%2F%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.nature.com%5C%2Fscitable%5C%2Fknowledge%5C%2Flibrary%5C%2Fdating-rocks-and-fossils-using-geologic-methods-107924044%5C%2F%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22webpage%22%2C%22title%22%3A%22Dating%20Rocks%20and%20Fossils%20Using%20Geologic%20Methods%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22Daniel%20J.%20Peppe%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22Alan%20L.%20Deino%22%7D%5D%2C%22abstractNote%22%3A%22Using%20relative%20and%20radiometric%20dating%20methods%2C%20geologists%20are%20able%20to%20answer%20the%20question%3A%20how%20old%20is%20this%20fossil%3F%22%2C%22date%22%3A%222013%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.nature.com%5C%2Fscitable%5C%2Fknowledge%5C%2Flibrary%5C%2Fdating-rocks-and-fossils-using-geologic-methods-107924044%5C%2F%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222025-12-27T21%3A27%3A53Z%22%7D%7D%5D%7D
anubha. (2024, November 11). Lab Work Made Easy: Choosing the Right Chemistry Glassware.
Scientific Lab Equipment Manufacturer and Supplier .
https://www.shivsons.com/lab-work-made-easy-choosing-the-right-chemistry-glassware/
The4. (2026, January 29).
A list of glassware equipment for high school chemistry lab. - Science . Science Equip.
https://www.scienceequip.com.au/blogs/news/a-list-of-glassware-equipment-for-high-school-chemistry-lab
says, G. johnson. (n.d.).
Beakers vs. Flasks: The Pros and Cons of Frequently Used Lab Glassware . Lab Pro Inc. Retrieved February 26, 2026, from
https://labproinc.com/blogs/laboratory-equipment/beakers-vs-flasks-the-pros-and-cons-of-frequently-used-lab-glassware
Understanding Laboratory Glassware Classifications . (n.d.). Globe Scientific. Retrieved February 26, 2026, from
https://globescientific.com/blogs/blog/understanding-laboratory-glassware-classifications
Laboratory Beakers Guide – Types & Uses Explained . (n.d.). ChemScience Inc. Retrieved February 26, 2026, from
https://www.chemscience.com/blog/www.chemscience.com
Lam, K., Armstrong, B., & Margelefsky, E. L. (2024). Correction to “Gravimetric and Thermal-Imaging Characterization of Water-Free Reflux Condensers for Laboratory Applications.”
ACS Omega ,
9 (20), 22508.
https://doi.org/10.1021/acsomega.4c03508
Lam, K., Armstrong, B., & Margelefsky, E. L. (2024). Gravimetric and Thermal-Imaging Characterization of Water-Free Reflux Condensers for Laboratory Applications.
ACS Omega ,
9 (14), 16563–16571.
https://doi.org/10.1021/acsomega.4c00411
Fordham, C. (2024, February 7). Condenser Chemistry: What’s the Difference Between Distillation & Reflux?
Asynt .
https://www.asynt.com/blog/condenser-chemistry-difference-between-distillation-reflux/
Fordham, C. (2024, February 7). Condenser Chemistry: What’s the Difference Between Distillation & Reflux?
Asynt .
https://www.asynt.com/blog/condenser-chemistry-difference-between-distillation-reflux/
admin. (2025, October 28). What Is Reflux Distillation? A Complete Guide.
Borosil Scientific .
https://www.borosilscientific.com/what-is-reflux-distillation-complete-guide/
Reflux and distillation. (n.d.).
Reflux and distillation – practical videos | 16–18 students . RSC Education. Retrieved February 18, 2026, from
https://edu.rsc.org/practical/reflux-and-distillation-practical-videos-16-18-students/4012294.article
Water flow in condenser - ECHEMI.com . (n.d.). ECHEMI. Retrieved February 18, 2026, from
https://www.echemi.com/community/water-flow-in-condenser_mjart2205112471_431.html
Abrahams, I., & Millar, R. (2008). Does Practical Work Really Work? A study of the effectiveness of practical work as a teaching and learning method in school science.
International Journal of Science Education ,
30 (14), 1945–1969.
https://doi.org/10.1080/09500690701749305
Vogel, A. I. (1977). A text-book of practical organic chemistry: incl. qualitative organic analysis (3. ed.; new impr). Longmans.
Indiana, W. Q. in. (2025, October 22).
Community Drinking Water Information - Madison County . Water Quality in Indiana.
https://www.in.gov/idem/cleanwater/drinking-water/community-drinking-water-info-madison-co/
Schneider, K. (n.d.).
These Indiana residents have brown water and want their town to tell them why . The Indianapolis Star. Retrieved February 5, 2026, from
https://www.indystar.com/story/news/environment/2025/10/06/indiana-brown-water-quality-safe-drinking/86481832007/
Fixed Costs vs Variable Costs: A Practical Guide with Examples . (n.d.). The Fino Partners. Retrieved February 5, 2026, from
https://thefinopartners.com/blogs/fixed-costs-vs-variable-costs-a-practical-guide-with-examples
Ohnstad, E. (2024, July 31). Can your business qualify for the R&D tax credit?
Abdo .
https://abdosolutions.com/can-your-business-qualify-for-the-rd-tax-credit/
Gibson, T. (2023, March 3). 5 Advantages of the Small Business R&D Tax Credit.
Omega Accounting Solutions .
https://omega-accounting.com/small-business-r-d-tax-credit/
Council, S. B. E. (2025, August 4). “One Big Beautiful Bill” Encourages Small Business Innovation: How Entrepreneurs Can Immediately Benefit from Restored R&D Expensing.
Small Business & Entrepreneurship Council .
https://sbecouncil.org/2025/08/04/one-big-beautiful-bill-encourages-small-business-innovation-how-entrepreneurs-can-immediately-benefit-from-restored-rd-expensing/
Water Quality Testing Business Plan Report 2026 | Setup . (n.d.). Retrieved February 5, 2026, from
https://www.imarcgroup.com/water-quality-testing-business-plan-project-report
Brockovich, E. (2025, November 12).
Who’s In Charge Of The Water In Madison County, Indiana? https://www.thebrockovichreport.com/p/whos-in-charge-of-the-water-in-madison
Amritha R. (2024, September 22).
THE IMPACT OF VARIABLE AND FIXED COST ON BUSINESS DECISION-MAKING | LinkedIn [Social Media]. Geojit Investments Limited. LinkedIn.
https://www.linkedin.com/pulse/impact-variable-fixed-cost-business-decision-making-amritha-r-3axbc/
365outsource_admin. (2025, October 24). How Small Businesses Cut Costs with Outsourcing.
365Outsource.Com .
https://www.365outsource.com/uncategorized/small-businesses-cut-costs-outsourcing/
Sullivan, P. (2023, September 19). Why You Need a Third-Party Assessment for Cybersecurity.
A-LIGN .
https://www.a-lign.com/articles/why-you-need-a-third-party-assessment-for-cybersecurity
What Is Third-Party Risk Management? (n.d.). Retrieved February 5, 2026, from
https://www.venminder.com/blog/what-is-third-party-risk-management
Water Quality Studies – Johnson Engineering . (n.d.). Retrieved February 5, 2026, from
https://johnsonengineering.com/services-projects/water-quality-studies/
Life Science Content Marketing Services. (n.d.).
Sciencia Consulting . Retrieved February 5, 2026, from
https://scienciaconsulting.com/content-marketing-for-life-sciences/
New Evidence That the R&D Tax Credit Promotes Research, Especially in Small Companies | ITIF . (n.d.). Retrieved February 5, 2026, from
https://itif.org/publications/2020/05/05/new-evidence-rd-tax-credit-promotes-research-especially-small-companies/
The Type of R&D That Small Businesses Can Really Use - NicheLabs, LLC. (2016, March 8).
NicheLabs .
https://www.nichelabs.com/general/the-type-of-rd-that-small-businesses-can-really-use/
The Type of R&D That Small Businesses Can Really Use - NicheLabs, LLC. (2016, March 8).
NicheLabs .
https://www.nichelabs.com/general/the-type-of-rd-that-small-businesses-can-really-use/
editor, P. R. F. B. P. R. is a copy, Economics, F.-C. with E. in, Finance, P., & policies, over twenty years of experience in the classroom L. about our editorial. (n.d.).
How Outsourcing Reduces Business Costs: Strategies and Examples . Investopedia. Retrieved February 5, 2026, from
https://www.investopedia.com/terms/o/outsourcing.asp
365outsource_admin. (2025, October 24). How Small Businesses Cut Costs with Outsourcing.
365Outsource.Com .
https://www.365outsource.com/uncategorized/small-businesses-cut-costs-outsourcing/
The Hidden Costs of R&D Outsourcing and How to Mitigate Them . (n.d.). Retrieved February 5, 2026, from
https://www.accelerance.com/blog/the-hidden-costs-of-outsourcing-rd-and-how-to-mitigate-them
Bob Kaiser. (2015, December 23). Simple machines [Edu].
KaiserScience .
https://kaiserscience.wordpress.com/physics/simple-machines/
shehlit. (2021, July 19). Student misconceptions about work and energy.
Chang Physics Class .
https://changphysicsclass.com/2021/07/19/student-misconceptions-about-work-and-energy/
Can you have “too much” mechanical advantage? (n.d.). Alpinesavvy. Retrieved January 29, 2026, from
https://www.alpinesavvy.com/blog/can-you-have-too-much-mechanical-advantage
Piatt, L. (2025, October 21). Understanding Pulley Systems for Mechanical Advantage.
Rigging Lab Academy .
https://rigginglabacademy.com/pulley-systems-for-mechanical-advantage/
Nave, C. R. (2017).
Simple Machines [Educational]. Georgia State University. HyperPhsyics.
http://hyperphysics.phy-astr.gsu.edu/hbase/Mechanics/simmac.html#c1
Kleynhans, L. (n.d.).
Unit 1: The mechanical advantage of an inclined plane . Retrieved January 28, 2026, from
http://ncvs3.books.nba.co.za/chapter/unit%e2%80%af1-the-mechanical-advantage-of-an-inclined-plane/
Mechanical Advantage Explained. (2017, April 2).
Educated Climber.Com .
https://www.educatedclimber.com/mechanical-advantage-explained/
Simple Machines: Mechanical Advantage | Research Starters | EBSCO Research . (n.d.). EBSCO. Retrieved January 28, 2026, from
https://www.ebsco.com
Urone, P. P., Hinrichs, R., Urone, P. P., & Hinrichs, R. (2020, March 26).
9.3 Simple Machines - Physics | OpenStax .
https://openstax.org/books/physics/pages/9-3-simple-machines
Simple Machines: Types, Activities, and Misconceptions . (n.d.). Studylib.Net. Retrieved January 28, 2026, from
https://studylib.net/doc/5444329/for-forces-and-motion-simple-machines-types-of-simple-mac...
Daniel J. Peppe, & Alan L. Deino. (2013).
Dating Rocks and Fossils Using Geologic Methods . Nature, Scitable.
https://www.nature.com/scitable/knowledge/library/dating-rocks-and-fossils-using-geologic-methods-107924044/